Charles Lee, PhD, FACMG, FRSC
Charles Lee is the Robert Alvine Family Endowed Chair, professor, and a board certified clinical cytogeneticist who has an active research program in the identification and characterization of structural genomic variants using advanced technology platforms. His laboratory was the first to describe genome-wide structural genomic variants (in the form of copy number variants (CNVs)) among humans[1] with the subsequent development of genomic maps[2][3] that are used in the diagnoses of array-based genetic tests. Lee served as the president of the Human Genome Organization (HUGO) from 2017 to 2023.Education
Lee attended Prairie River Junior High School in High Prairie, Alberta, and later McNally Composite High School in Edmonton. He completed all of his postsecondary education at the University of Alberta, earning a BSc in Genetics (1990), an MSc in Experimental Pathology (1993), and a PhD in Medical Sciences (1996).
For his MSc thesis, titled "Tandemly Repetitive DNA in the Karyotypic and Phylogenetic Evolution of the Cervidae Family," Lee identified interstitial telomeric DNA sequences in the giant chromosomes of the Indian muntjac (Muntiacus muntjak vaginalis) [4]. These sequences are thought to be remnants left after the rapid fusion of ancestral acrocentric chromosomes that gave rise to the species' present-day giant chromosomes.
His PhD thesis, "Identification and Characterization of Two Mammalian Centromeric Satellite DNA Families," described the discovery of human gamma satellite DNA and cervid satellite I DNA. Lee's mentor for both graduate degrees was Dr. Chyi Chyang (C.C.) Lin, a clinical and mammalian cytogeneticist. His graduate advisory committee included Drs. John Kuspira, Bruce Ritchie, Jerome B. Rattner, Gary D. Burkholder, Patrick Ferreira, and Robert A. Stinson.
Post Doctoral Training
After completing his PhD, Lee conducted postdoctoral research in molecular cytogenetics with Professor Malcolm A. Ferguson-Smith at the University of Cambridge, UK, supported by a Natural Sciences and Engineering Research Council of Canada Fellowship (1996-1998). He then pursued clinical cytogenetics training with Dr. Cynthia C. Morton at Brigham and Women's Hospital and Harvard Medical School (1998-2001), subsequently earning certification from the American Board of Medical Genetics and being named a Fellow of the American College of Medical Genetics (FACMG).

Research
Discovery of widespread structural variation in the human genome
In 2001, Lee established his research laboratory at Brigham and Women's Hospital / Harvard Medical School, first located on the 6th floor of the Thorn Building and later moving to the 4th floor of the Eugene Braunwald Research Center at 221 Longwood Avenue. In 2003, his team began using array-based comparative genomic hybridization and unexpectedly detected widespread copy number variation (CNVs) in the genomes of 55 healthy individuals. The findings were published online in Nature Genetics on August 1, 2004[1]. Around the same time, Sebat et al. reported similar observations in the July 23, 2004 issue of Science. These two studies have since been widely cited as landmark contributions that defined the field[5][6]. The significance of this discovery—and the scientific community's response—was further recognized when Science named "human genetic variation” the 2007 Breakthrough of the Year[7].

Structural genomic variation in model organisms and evolution
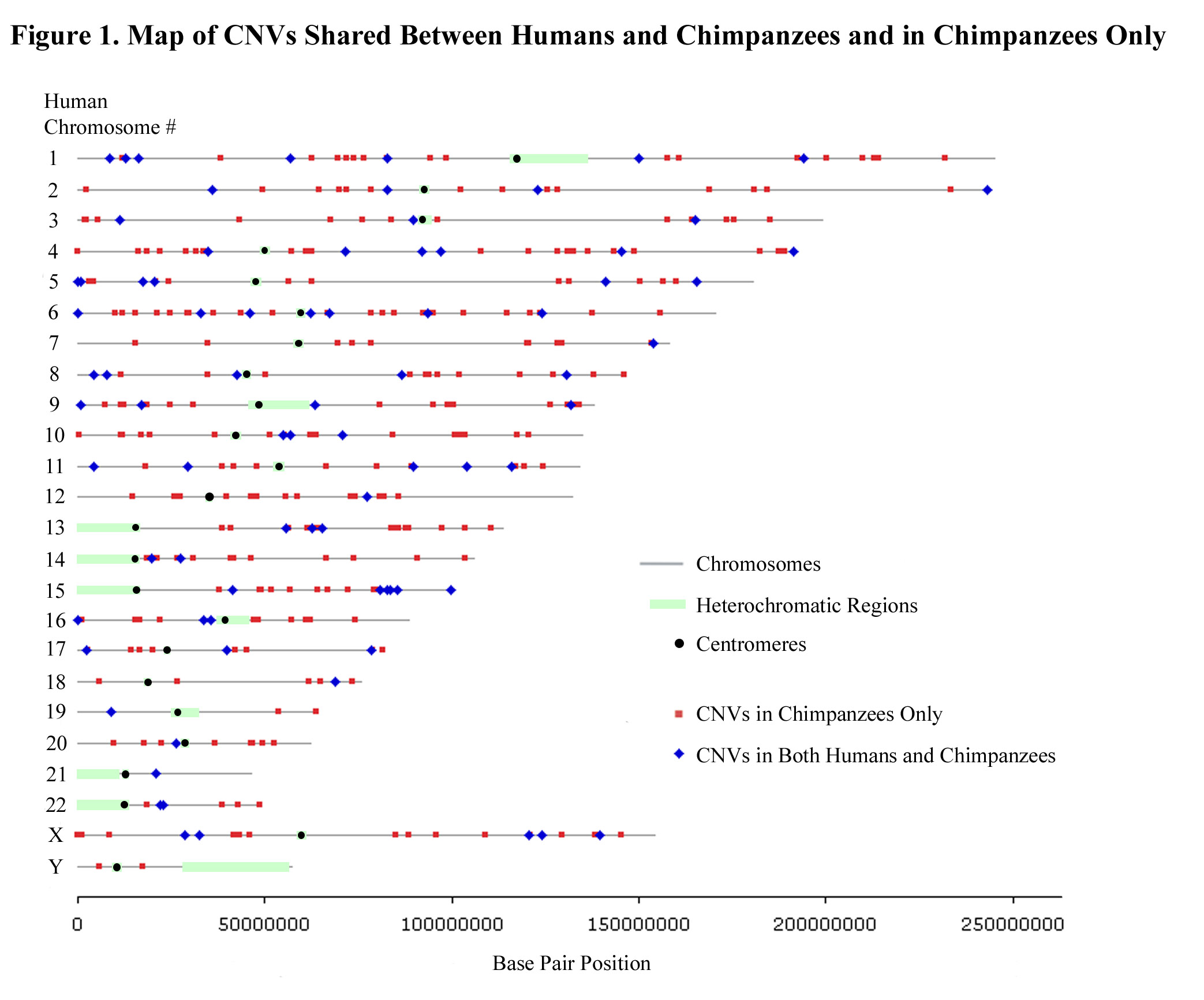
One of Lee's long-standing scientific interests is understanding how structural genomic variation shapes the phenotypes of model organisms and contributes to chromosomal and organismal evolution. His MSc work clarified the tandem chromosome fusion events that produced the giant chromosomes of the Indian muntjac deer (2n = 6 in females, 2n = 7 in males), the mammal with the lowest diploid chromosome number[8].
Following his discovery of widespread structural variation in the human genome, Lee generated the first copy number variation (CNV) maps for the chimpanzee [9],[10], rhesus macque[11], and zebrafish[12].
In 2007, Lee and colleagues reported that increased copy number of the salivary amylase gene AMY1—which enhances oral starch digestion—likely provided a selective advantage to human populations with high-starch diets[13]. This was the first study to show that CNVs can be subject to positive selection in response to cultural shifts, such as the development of agriculture, and it has become a well known example of the evolutionary significance of CNVs — now a standard topic in human genetics textbooks. More recently, Lee's group used molecular archaeology to show that an initial duplication of AMY1 was present in the genomes of three Neanderthals and one Denisovan[14], suggesting that the first extra copy arose as early as 800,000 years ago, before the divergence of Neanderthals and modern humans.
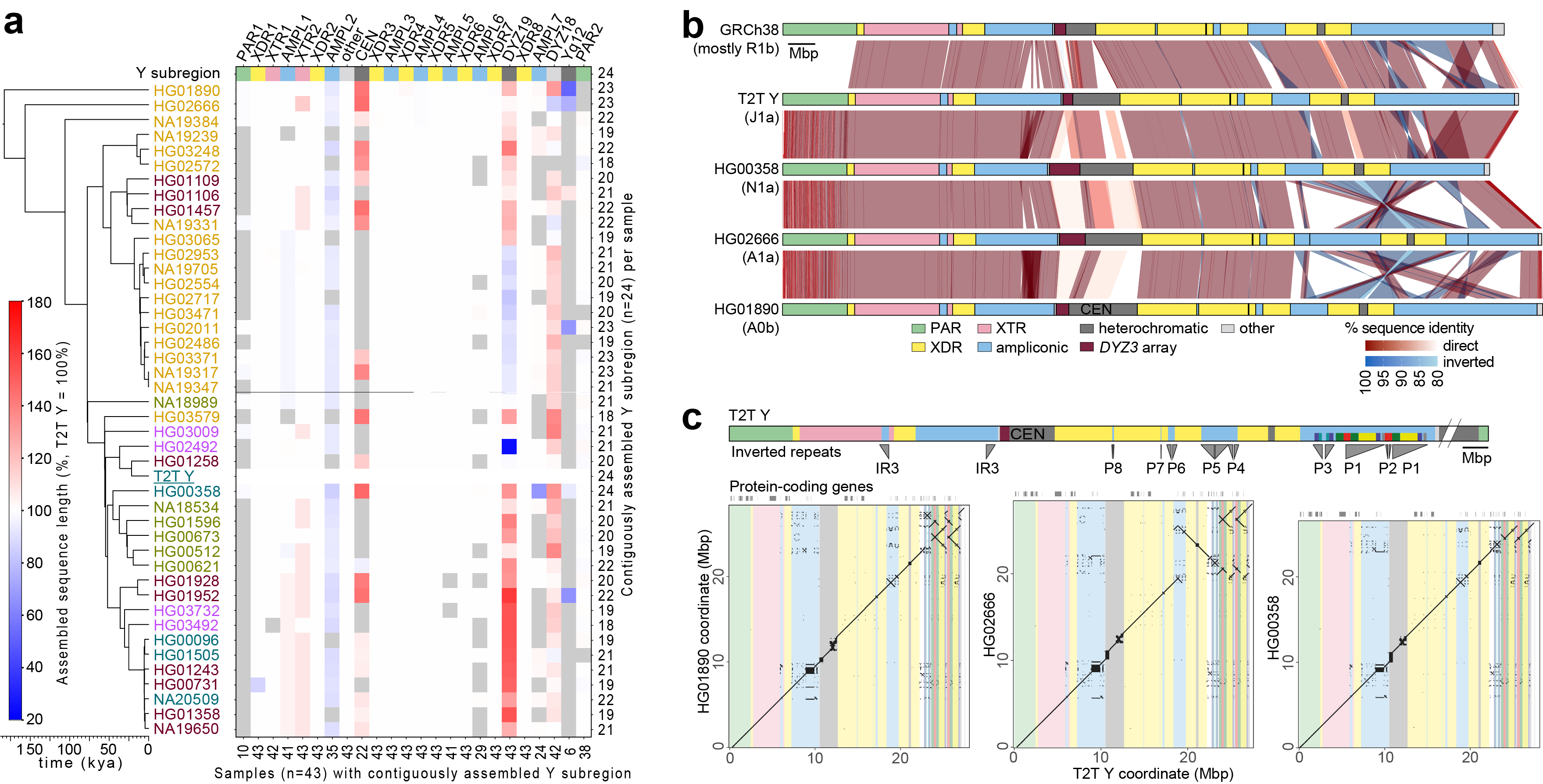
In 2023, Lee and colleagues produced one of the first two comprehensive sequence reports [15],[16] of the human Y chromosome. His team analyzed the Y chromosomes of 43 genetically diverse males, uncovering extensive structural variation with important implications for research on male infertility, cancer immunology, and cardiovascular disease.
New technologies and algorithms for structural genomic variation detection
After generating the first copy number variation map of the human genome [2], Lee co-led the Structural Variation Group of the International 1000 Genomes Project from 2008 to 2015, working alongside Jan O. Korbel, Evan E. Eichler, and Matthew E. Hurles. He later helped found the Human Genome Structural Variation Consortium (https://www.hgsvc.org), which he now co-leads with Jan O. Korbel, Evan E. Eichler, and Tobias Marschall.

LEADERSHIP
As the inaugural director of The Jackson Laboratory for Genomic Medicine (2013-2024), Lee oversaw the creation of a new 185,000-square-foot, stand-alone institute housing a state-of-the-art genomics research center dedicated to uncovering the complex causes of human disease and developing new diagnostics and therapeutics.
During his time with The Jackson Laboratory for Genomic Medicine, Lee also served as a faculty member at different universities in Asia: First at Seoul National University College of Medicine (2013-2015), then at Ewha Womans University (2015-2020), and then finally at the Xi'an Jiaotong University First Affiliated Hospital (2018-2022). While at Ewha Womans University, he established a scholarship program that would permit up to ten students each year to attend the McKusick Short Course for Medical and Mammalian Genetics.
In March 2020, Dr. Lee led The Jackson Laboratory (JAX) in rapidly launching COVID-19 diagnostic testing, immediately expanding the state of Connecticut's testing capacity to 10,000 tests per day. He framed this effort as a humanitarian responsibility and in April 2020, Governor Lamont appointed Dr. Lee as one of eight experts on the Reopen Connecticut Advisory Task Force, where he provided scientific guidance on safely reopening the state's economy and public institutions.
Dr. Lee's leadership and his expertise were central to Connecticut's public health strategy during the pandemic, and he was subsequently recognized by the Hartford Business Journal as one of the top 25 leaders who significantly impacted the state's COVID-19 response.
On an international level, Lee served as president of the Human Genome Organization (HUGO) from 2017-2023, during which he led the successful merger and integration of the Human Genome Variation Society (HGVS) and the Human Variome Project into HUGO.
Awards and Honors
In 2008, Lee received the prestigious Ho-Am Prize in Medicine — becoming the youngest recipient in the award's history at age 39. He was honored "for contributions to the discovery of copy number variants (CNVs) in human genetics and the creation of a new map of human genetic variation, paving the way for major advances in the field.”
In 2014, he was named a Thomson Reuters Citation Laureate "for the discovery of large-scale copy number variation and its association with disease." In announcing the honor, Thomson Reuters highlighted the exceptional global impact of his work, citing its extraordinarily high citation record and its significance as the type of discovery considered worthy of Nobel recognition.
Lee has also been celebrated by his alma mater, the University of Alberta, receiving its highest alumni honor — the Distinguished Alumni Award (2018) — and later an honorary Doctor of Science degree (2024) "in recognition of the groundbreaking discovery of structural variation in the human genome and his sustained advocacy for science and lifelong learning."
Lee is a Fellow of the American Association for the Advancement of Science (2012), the Korean Academy of Science and Technology (2024). In 2024, Lee became a Fellow of The Royal Society of Canada (Division of Medical Sciences). His candidature reads:
"Charles Lee made a revolutionary discovery in 2004: genes don't always occur in two copies per cell. This insight transformed human genetics, reshaping our understanding of disease mechanisms and evolution, and enabling new diagnostic technologies.”
References
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004 Sep;36(9):949-51.
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006 Nov 23;444(7118):444-54.
- Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010 Apr 1;464(7289):704-12.
- Lee, C. (1993). Tandemly Repetitive DNA in the Karyotypic and Phylogenetic Evolution of the Cervidae Family. MSc Thesis, University of Alberta.
- Finding the right lenses. Nat Genet 39 (Suppl 7), S1 (2007)
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004 Jul 23;305(5683):525-8.
- Pennisi E (2007) Human Genetic Variation. Science 318, 1842-1843
- Lee C, Sasi R, Lin CC. Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs. Cytogenet Cell Genet. 1993;63(3):156-9. doi: 10.1159/000133525. PMID: 8485991.
- Perry GH et al. (2006). Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA. 103, 8006-11.
- Perry GH et al. (2008). Copy number variation and evolution in humans and chimpanzees. Genome Res. 18, 1698-710.
- Lee AS et al. (2008). Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 17, 1127-36.
- Freeman JL et al. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genomics. 2007 Jun 27; 8: 195
- Perry, G.H. et al. Diet and the evolution of human amylase gene copy number variation. Nature Genetics 39, 1256-1260 (2007).
- Yilmaz F, et al. (2024). Reconstruction of the human amylase locus reveals ancient duplications seeding modern-day variation. Science Oct 17: eadn0609.
- Hallast, P., Ebert, P., Loftus, M. et al. (2023) Assembly of 43 human Y chromosomes reveals extensive complexity and variation. Nature 621, 355-364.
- Rhie, A., Nurk, S., Cechova, M. et al. (2023) The complete sequence of a human Y chromosome. Nature 621, 344-354.